Four of our papers have been accepted by MICCAi2025
I am delighted to share four of our papers have been accepted by MICCAI2025
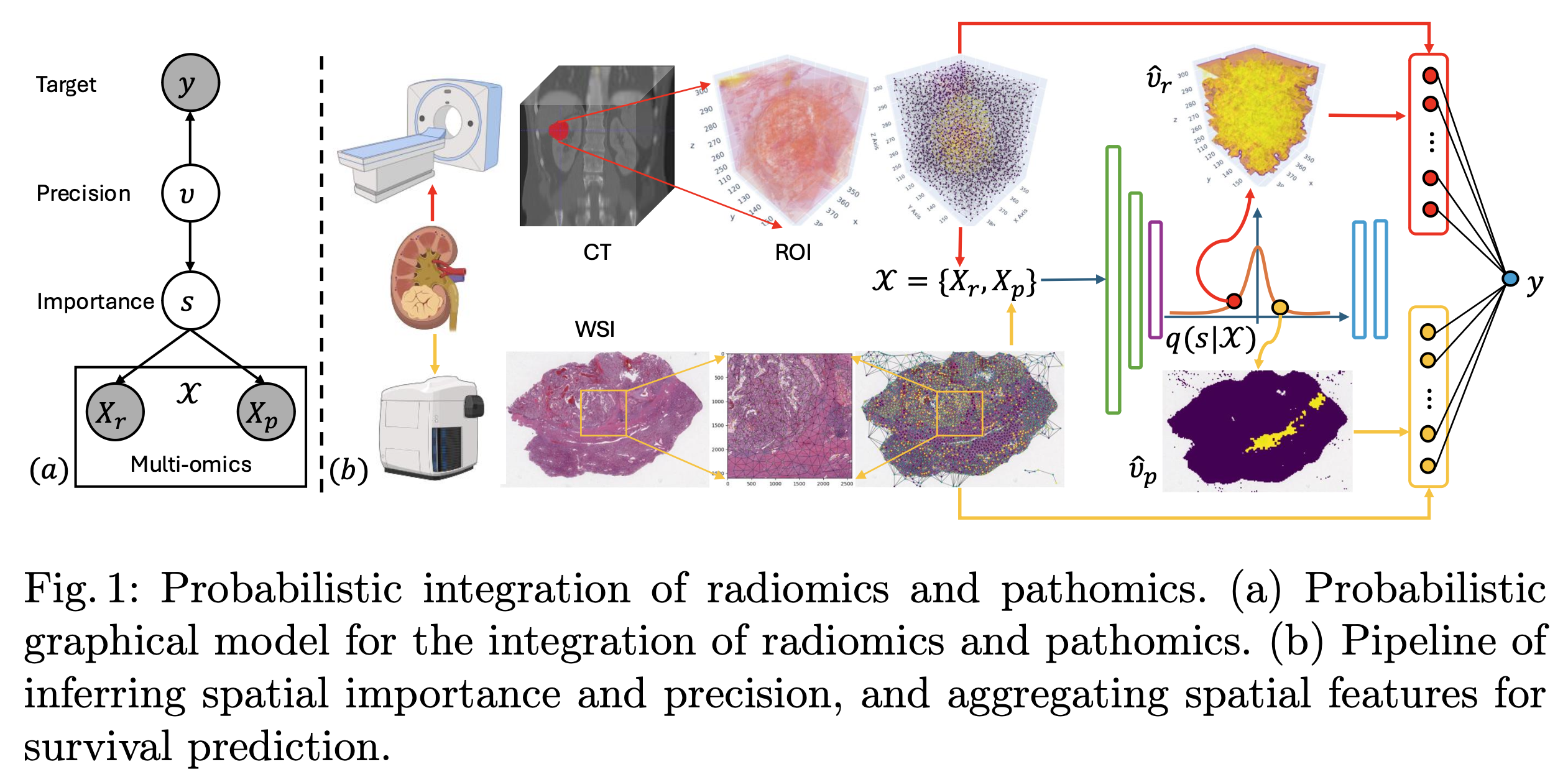
Multimodal data integration
S. Gao et al., Probabilistic Integration of Renal Cancer Radiology and Pathology Using Graph Neural Networks
Abstract. Kidney tumors can be highly heterogeneous from the microscopic to the macroscopic scale. To address this, we propose a sparsity-informed probabilistic integration of radiomics and pathomics for kidney cancer analysis. We construct radiology and pathology graphs to model spatial correlations, then use a probabilistic method and graph neural networks to identify biomarkers and aggregate spatial features. Our validation shows that this integrated approach significantly outperforms traditional methods in kidney survival analysis, achieving a notable improvement of 0.084 in the concordance index, enabling better prognostic assessments for kidney cancer patients.
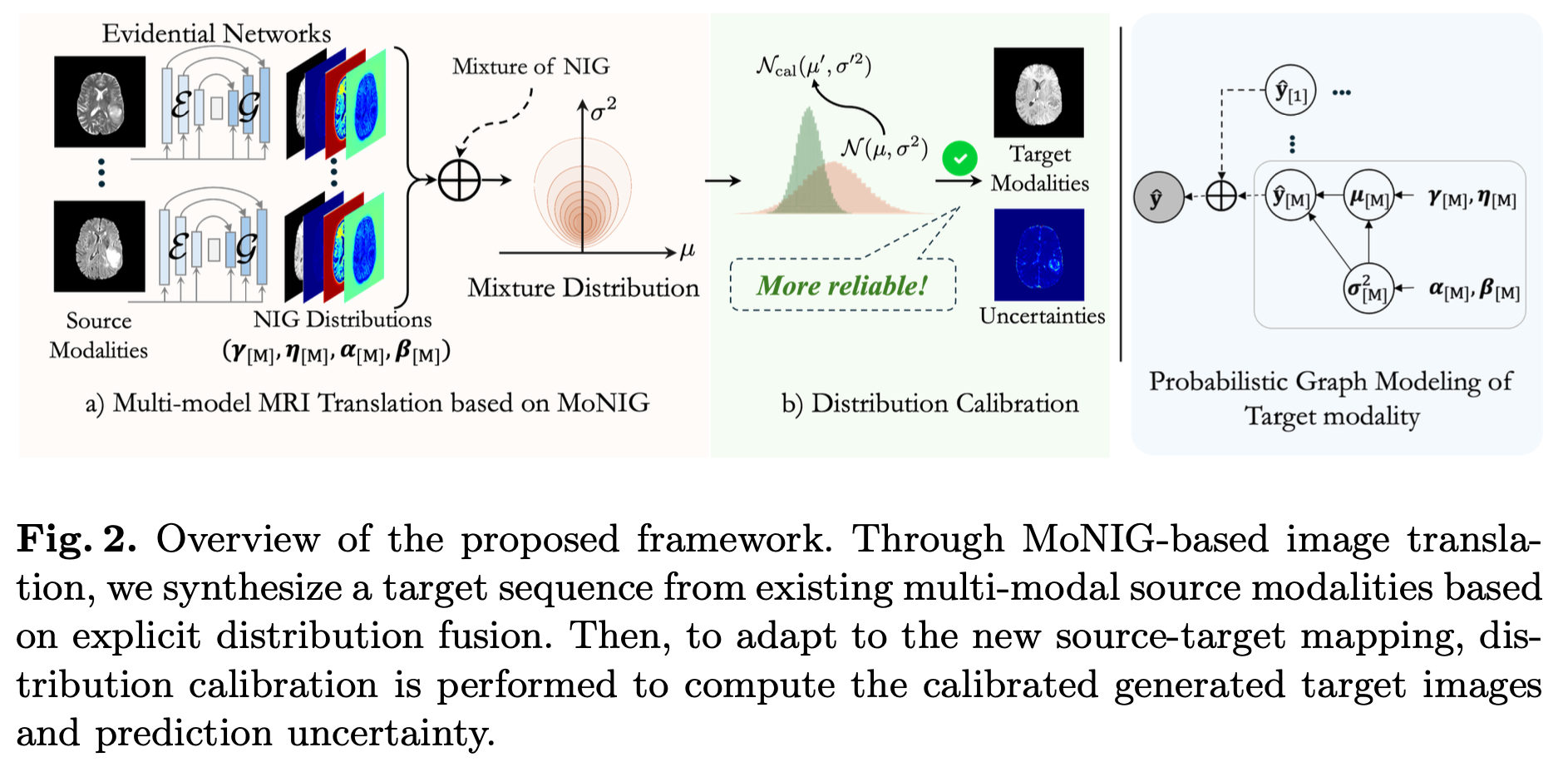
Multimodal MRI translation
J. Liu et al., Multi-modal MRI Translation via Evidential Regression and Distribution Calibration
Abstract. Multi-modal Magnetic Resonance Imaging (MRI) translation leverages information from source MRI sequences to generate target modalities, enabling comprehensive diagnosis while overcoming the limitations of acquiring all sequences. While existing deep-learning-based multi-modal MRI translation methods have shown promising potential, they still face two key challenges: 1. lack of reliable uncertainty quantification for synthesized images, and 2. limited robustness when deployed across different medical centers. To address these challenges, we propose a novel framework that reformulates multi-modal MRI translation as a multi-modal evidential regression problem with distribution calibration. Our approach incorporates two key components: 1. an evidential regression module that estimates uncertainties from different source modalities and an explicit distribution mixture strategy for transparent multi-modal fusion, and 2. a distribution calibration mechanism that adapts to source-target mapping shifts to ensure consistent performance across different medical centers. Extensive experiments on three datasets from the BraTS2023 challenge demonstrate that our framework achieves superior performance and robustness across domains.
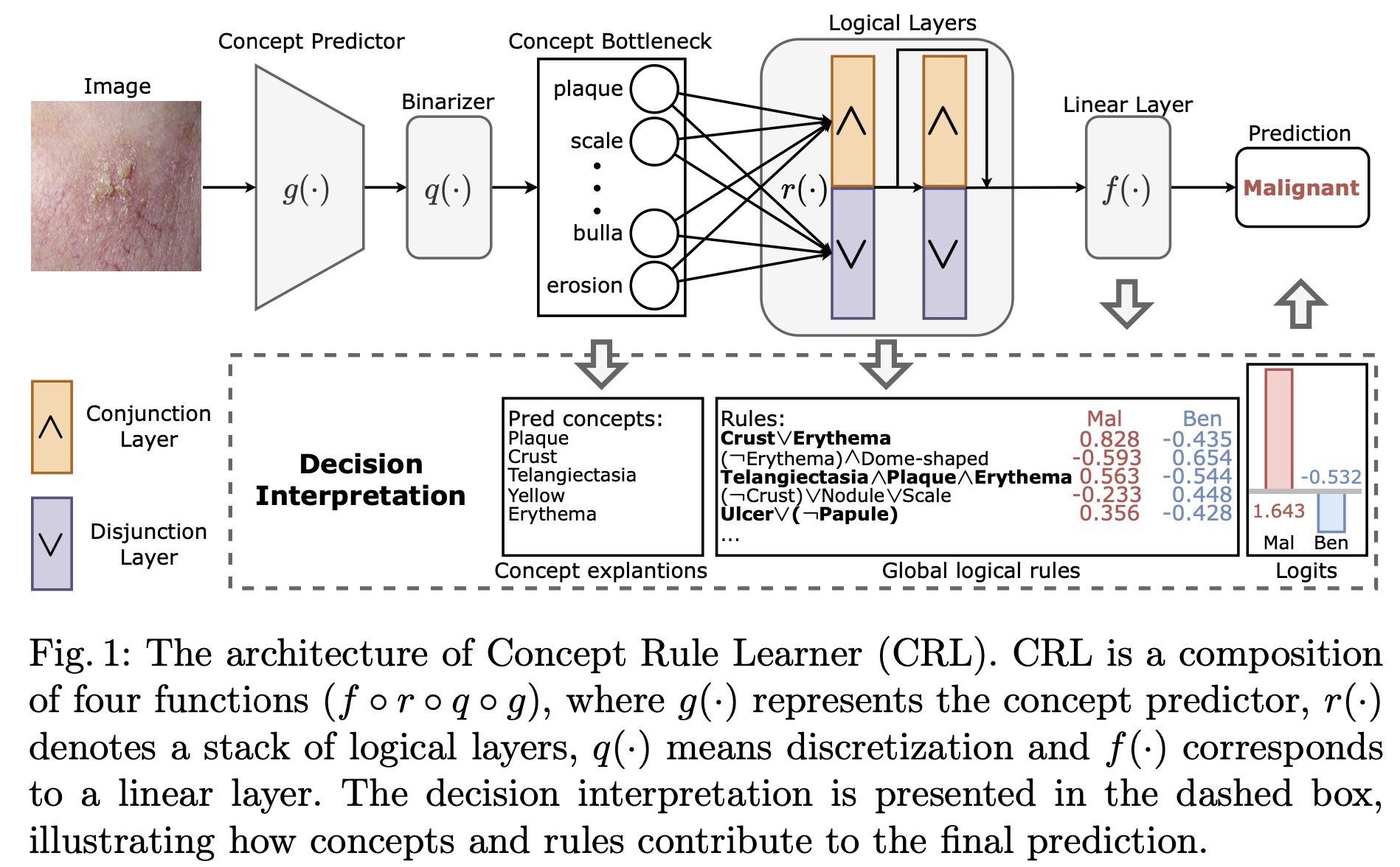
Interpretable and generalizable medical image classfication
Y. Gao et al., Learning Concept-Driven Logical Rules for Interpretable and Generalizable Medical Image Classification
Abstract. The pursuit of decision safety in clinical applications highlights the potential of transparent methods in medical imaging. While concept-based models offer local concept explanations (instance-level), they often neglect the global decision logic (dataset-level). Moreover, these models often suffer from concept leakage, where unintended information within soft concept representations undermines both interpretability and generalizability. To address these limitations, we propose Concept Rule Learner (CRL), a novel framework to learn Boolean logical rules from binary visual concepts. CRL employs logical layers to capture concept correlations and extract clinically meaningful rules, thereby providing both local and global interpretability. The results from two tasks demonstrate that CRL achieves competitive performance with existing interpretable methods while improving generalizability to out-of-distribution data. The code of our work is available anonymously at https://anonymous.4open.science/r/crl-73E4.
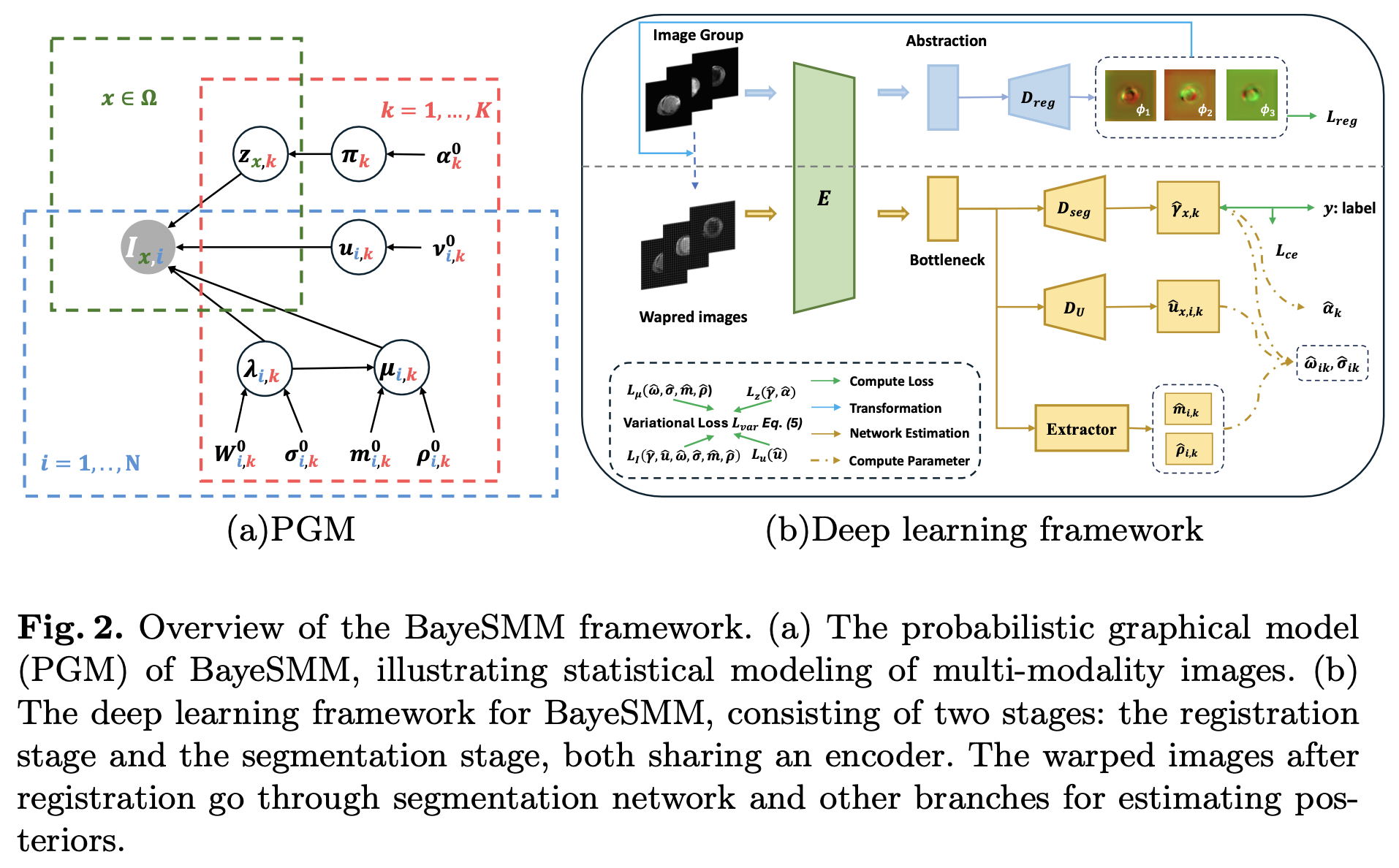
Combined image registration and segmentation
Y. Liu et al., BayeSMM: Robust Deep Combined Computing Tackling Heavy-tailed distribution in Medical Images
Abstract. Abnormal structures in multi-modality medical images often lead to heterogeneous heavy-tailed distributions. However, traditional models, especially those relying on Gaussian distributions, struggle to effectively capture these outliers. To address this, we propose BayeSMM, a novel framework that leverages Student’s t distribution mixture models (SMM) to simultaneously perform registration and segmentation for misaligned multi-modality medical images. Specifically, we construct a Bayesian Student’s t mixture model incorporating the heavy-tailed nature of the Student’s t distribution and develop variational inference to optimize the model. Guided by variational inference, we design a novel deep learning architecture that performs registration and segmentation jointly. We demonstrate the effectiveness of BayeSMM with experiments on the MS-CMR dataset, where the results show superior performance compared to existing combined computing methods, and yield enhanced robustness under the simulated heavy-tailed setting.